On August 28th, 2018, Professor Heng-Yu Fan’s group published a paper entitled “CFP1 Coordinates Histone H3 Lysine-4Trimethylation and Meiotic Cell Cycle Progressionin Mouse Oocytes” in Nature Communications.
Several research groups have demonstrated that the amount of H3K4me3 remarkably increases during oocyte maturation, and H3K4me3 is abundantly deposited at numerous broad regions on chromatin of mature oocytes, but the functional significance of these phenomena remain unclear.A recent study by Fan laboratory (Yu et al, Cell Reports 2017) indicated that the CFP1-SETD1 complexis the predominant H3K4 methyltransferases in most species and cell types including mouse oocytes.Conditional knockouts of Cxxc1 (the gene coding for CFP1) or Setd1b in oocytes caused oocyte maturation defects and female infertility, but the mechanism was investigated insufficiently.
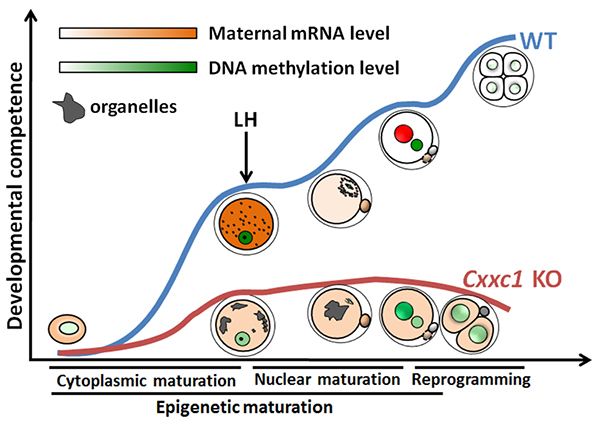
A diagram showing role of maternal CFP1 in the establishment of developmental competence during oocyte-to-embryo transition
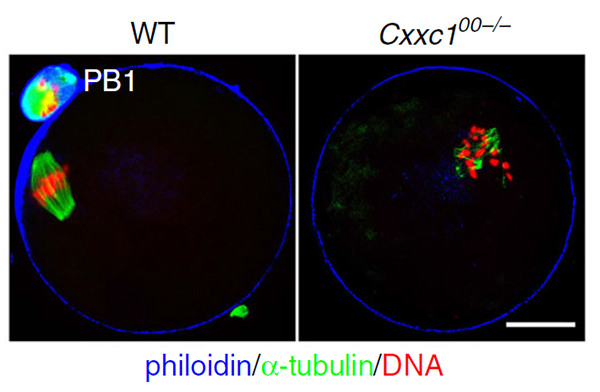
Cxxc1-deleted oocytes have defects of meiotic maturation
SETD1�CFP1 complex is dually inhibited by CDK1-triggered CFP1 phosphorylation and degradation at the onset of the G2�M transition. Preventing CFP1 degradation and phosphorylation in oocytes and zygotes caused CFP1 accumulation on condensed chromosomes particularly in the centromeric region and additively impaired meiotic maturation and preimplantation embryo development, respectively. Thus, the downregulation of H3K4me3 and removal of CFP1 from chromatin are crucial for proper chromatin structure that is required for cell cycle progression.
CFP1 protein is transiently degraded during not only the meiotic division of mouse oocytes, but also the mitotic divisions of early embryo blastomeres and HeLa cells.The finding of complete degradation of CFP1 during each cell division reveals a surprising fact that all SETD1�CFP1 H3K4 methyltransferase complexes need to be de novo assembled in the cell after each division, and the distribution pattern of this complex on chromatin needs to be re-established in every cell cycle. This knowledge is conceptually important for understanding the genome-wide regulation of H3K4 methylation dynamics in general.
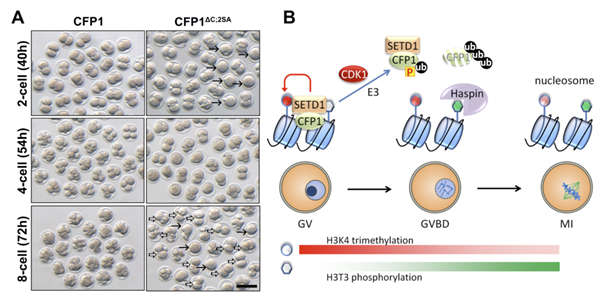
A:Expression of constitutively active CFP1 causes developmental abnormalities in preimplantation embryos.
B:CFP1is dual regulated by polyubiquitination and phosphorylation in meiotic cell cycle.
Links: https://www.nature.com/articles/s41467-018-05930-x