On Dec. 16, 2019, Dr. Xing Guo’s laboratory from Life Sciences Institute of Zhejiang University, published a research article in PNAS entitled “Reversible phosphorylation of Rpn1 regulates 26S proteasome assembly and function”. For the first time, the mechanism of a phosphosite of proteasome subunit regulates proteasomal precursor assembly and function was explored.
Background:
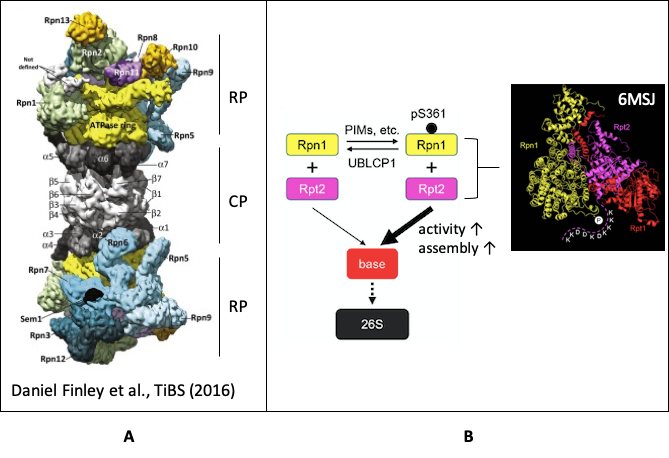
The ubiquitin proteasome system (UPS) is responsible for the degradation of misfolded, damaged, or unneeded cellular proteins. This pathway is important for the maintenance of cell cycle progression, DNA repair, apoptosis, stress response and protein homostasis for eukaryotes. Function defected proteasome related to several diseases, such as multiple myeloma, aging and alzheimer's disease. The 26S proteasome consists of the 20S core particle (CP) and the 19S regulatory particle (RP). The CP is a barrel-shaped complex responses for protein degradation. The RP mediates substrate unfolding, CP gate opening, and translocation of substrates into the CP (Fig A). On the other hand, the proteasome has been shown to be regulated by multiple posttranslational modifications, one of the most common is phosphorylation. Rpn1 of the 19S base is a receptor of ubiquitin and proteins with ubiquitin-like (UBL) domains. The Ser361 on Rpn1 is among the top 10 most frequently detected human proteasome phosphosites by mass spectrometry that are conserved in vertebrates. However, the function of this phosphosite is unknown.
Abstract:
The fundamental importance of the 26S proteasome in health and disease suggests that its function must be finely controlled, and yet our knowledge about proteasome regulation remains limited. Posttranslational modifications, especially phosphorylation, of proteasome subunits have been shown to impact proteasome function through different mechanisms, although the vast majority of proteasome phosphorylation events have not been studied. Here, we have characterized 1 of the most frequently detected proteasome phosphosites, namely Ser361 of Rpn1, a base subunit of the 19S regulatory particle. Using a variety of approaches including CRISPR/Cas9-mediated gene editing and quantitative mass spectrometry, we found that loss of Rpn1-S361 phosphorylation reduces proteasome activity, impairs cell proliferation, and causes oxidative stress as well as mitochondrial dysfunction. A screen of the human kinome identified several kinases including PIM1/2/3 that catalyze S361 phosphorylation, while its level is reversibly controlled by the proteasome-resident phosphatase, UBLCP1. Mechanistically, Rpn1-S361 phosphorylation is required for proper assembly of the 26S proteasome, and we have utilized a genetic code expansion system to directly demonstrate that S361-phosphorylated Rpn1 more readily forms a precursor complex with Rpt2, 1 of the first steps of 19S base assembly (Fig B). These findings have revealed a prevalent and biologically important mechanism governing proteasome formation and function. The discover also may lead to new approaches of manipulating proteasome activity for therapeutic purposes.
Ph.D. candidate Xiaoyan Liu in Guo lab and Weidi Xiao in Ping Xu lab of Beijing Proteome Research Center are co-first authors. Xing guo is the corresponding author. This research was supported by Natural Science Foundation of China, Zhejiang Natural Science Foundation, Fundamental Research Funds for the Central Universities and the startup funding from Zhejiang University.
Link: https://www.pnas.org/content/early/2019/12/13/1912531117/tab-figures-data