Dong Fang’s laboratory published a research article entitled Joint single-cell multiomic analysis in Wnt3a induced asymmetric stem cell division in Nature Communications on October 12th,2021. The authors established a new sequencing method: SET-seq (same cell epigenome and transcriptome sequencing), which could jointly profile the epigenome and transcriptome in the same single cell. Utilizing this method, the authors investigated the Wnt3a induced mouse embryonic stem cells asymmetric division, and provided mechanistic insights into the gene regulatory programs for maintaining and resetting stem cell fate during differentiation.
Wnt signaling plays an important role in stem cell maintenance, tumorigenesis, and tumor metastasis. Wnt signaling usually functions through the spatial distribution of WNT ligands and thus in vivo studies of Wnt signaling are difficult. Previous studies have shown that WNT3a proteins coated beads provide a spatially restricted Wnt signaling, and can induce asymmetric cell division of mouse embryonic stem cells (mESCs). When one mESC is attached to Wnt3a beads and divides after one cell cycle, the daughter cell proximal to Wnt3a beads maintains cellular pluripotency and the distal daughter cell exhibits a progressive differentiation status1. In this process, the parental and newly synthesized histones are asymmetrically segregated into proximal and distal cells, resulting in differential inheritance of histones in two daughter cells2. However, the detailed transcriptional and epigenomic changes underlying this asymmetric cell division have not been elucidated.
In the SET-seq, the nuclei and cytoplasmic RNA are separated from the same cells. Then, the epigenetic library is constructed by Tn5 transposomes to profile the chromatin binding proteins in the nuclei. In the meantime, cDNA is reversely transcribed from the cytoplasmic RNA for the transcriptome library construction (Fig. 1A). The required number of cells for the SET-seq can be very low, and even a single cell can be used. In addition, it’s easy to adopt this method in the laboratory as the libraries are easy to construct. The authors jointly profile H3K27me3 and H3K4me3 along with gene expression in Wnt3a inducted mESC asymmetric division. The daughter cells are clustered into the Mix, Proximal and Distal clusters based on their gene expression profiles. In addition, the cells in the Proximal and Distal clusters are enriched with asymmetric divided cells and cells in the Mix cluster are symmetrically divided. When compared with gene expressions, H3K27me3, but not H3K4me3, is highly correlated with gene expressions in the asymmetric cell division (Fig. 1B). Moreover, H3K27me3 is enriched at the marker genes of the ‘Mix’ clusters, suggesting that H3K27me3 is the key regulator for the asymmetric cell division. In the subunits of PRC2 which is the methyltransferase complex for H3K27me3, only Aebp2 is elevated in the Mix cluster containing symmetrically divided cells. Knockout of Aebp2 increases the ratio of asymmetrically divided cells. These results further confirm that H3K27me3 is the key regulator for Wnt3a induced asymmetric cell division.
Dr. Dong Fang is the corresponding author of this paper. Zhongxing Sun, Yin Tang, Yanjun Zhang and Yuan Fang are the co-first authors. This work is supported by the National Natural Science Foundation of China.
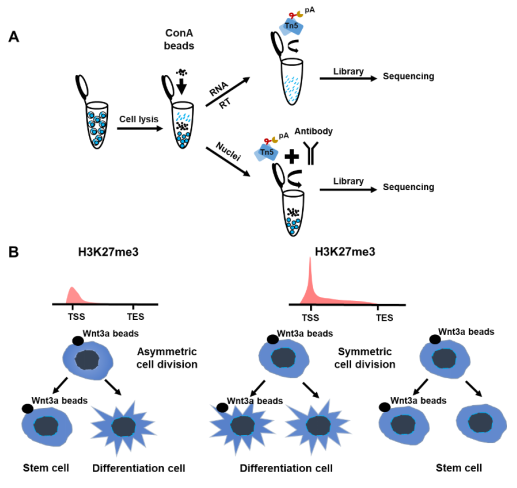
Figure 1. H3K27me3 as key regulator in Wnt3a induced asymmetric cell division. A. The workflow of SET-seq. ConA beads: Concanavalin A beads. B. H3K27me3 plays an important role in asymmetric cell division. TSS, Transcription Start Site; TES, Transcription End Site.
https://doi.org/10.1038/s41467-021-26203-0