On July 11, 2023, Shixian Lin’s Lab in our institute published a research article in JACS entitled “Manipulating cation-π interactions of reader proteins in living cells with genetic code expansion”.
Cation-π interactions are among the strongest non-covalent interactions between the cation and the π system, and are recognized by the chemistry, biology, and material science communities as a prominent facet of molecular recognition. Cation-π interactions play a central role in a wide variety of biological processes, including molecular recognition, adhesion, folding, and biological self-assembly.
Despite tremendous success in understanding the chemical nature and the importance of cation-π interactions in a range of biological processes, particularly in epigenetic regulation, the design and synthesis of stronger cation-π interactions in living cells remain largely elusive. In this paper, the authors design several electron-rich Trp derivatives and incorporate them into histone methylation reader domains to enhance the affinity of the reader domains for histone methylation marks via cation-π interactions in living cells. The authors show that this site-specific Trp replacement strategy is generally applicable for the engineering of high-affinity reader domains for the major histone H3 trimethylation marks such as H3K4me3, H3K9me3, H3K27me3, and H3K36me3, with high specificity. Furthermore, the authors demonstrate that engineered reader domains can serve as powerful tools for the enrichment and imaging of histone methylation, as well as for capturing the protein interactome at chromatin marks in living cells. Therefore, this approach paves the way for the design of enhanced cation-π interactions in reader proteins in living cells for various biological applications.
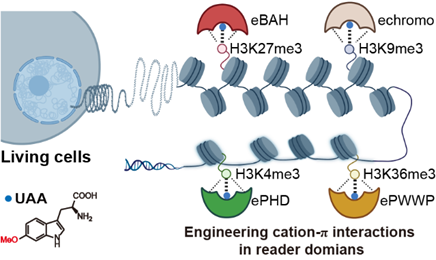
Figure 1. Engineering cation-π interactions of histone H3 trimethylation readers with high affinity and specificity in living cells via genetic code expansion.
Dr. Hongxia Zhao, Ling Tang and Yu Fang are co-first authors in this study. Dr. Shixian Lin is the corresponding author. This research was supported by the National Key R&D Program of China (Grant 2019YFA09006600), the National Natural Science Foundation of China (Grant 92253302, 22222705, 22207096, 91953113), and China Postdoctoral Science Foundation (Grant 2022M712782).
Link:https://pubs.acs.org/doi/10.1021/jacs.3c02293