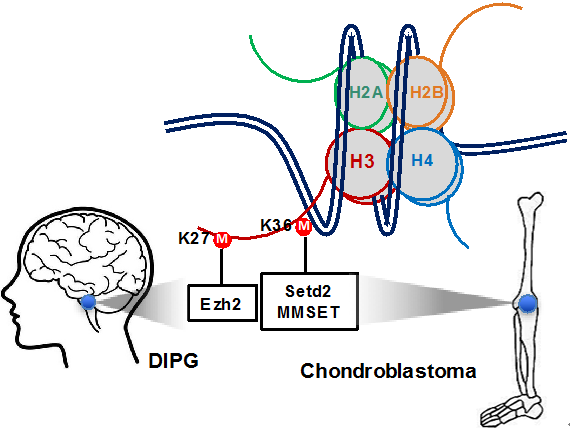
Fig 1. Oncohistone H3.3K27M and H3.3K36M altered chromatin states in DIPG and chondroblastoma cases respectively
Recent genetic studies have identified two somatic oncohistone mutations, H3.3K27M and H3.3K36M, in over 75% DIPG cases and in more than 90% chondroblastoma cases, respectively. With the high incidence of histone H3 lysine to methionine mutations, it is critically important to understand how these methionine substitutions promote tumorigenesis.We have shown that oncohistone H3.3K27M reprograms H3K27 methylation and gene expression in DIPG cases, as well as oncohistone H3.3K36M inhibits H3K36 methyltransferases to reprograms the epigenome of chondroblastomas (Fig. 1). We will use biochemistry, molecular biology, epigenomic approaches, and large screening to study the function of oncohistone mutations and the molecular mechanisms for the inheritance of epigenetic information.
Aim 1: Determine to what extent oncohistones promote the formation of tumors.
Our initial studies have provided fundamental information about the molecular mechanisms of how oncohistones, H3.3K27M and H3.3K36M, drive tumor formation. However, the detailed mechanism of oncohistone mutation induced tumorigenesis is still largely unknown. We will systematically study the impacts of different oncohistone mutations in distinct cell types to elucidate the function of oncohistones.
Aim 2: Screen the potential drug targets for tumors harboring oncohistone.
Since the incidence of oncohistone mutations is high in tumors, we will use large screening to find novel drug targets in these tumors to fight tumors.
Aim 3: Determine how epigenetic information is precisely distributed
Besides the DNA sequence, epigenetic information has to be inherited during cell cycle. In addition, histone modifications defects have been found in different tumors. it’s important to understand how epigenetic information is precisely distributed across the genome. We will use biochemistry, molecular biology, cell biology and epigenomic approaches to study:
3.1 How histone methylation is precisely distributed across the genome
3.2 The interaction among different epigenetic modification