Mechanism of action of ribosome-targeting antibiotics
The emergence of antibiotic-resistant bacteria has created a pressing need to understand the structural basis of the action of antibiotics that is essential in developing improved antimicrobials and instrumental in elucidating the mechanisms of cellular processes.Bacterial protein synthesis is a major target for many antibiotics that act upon alternating messenger RNA(mRNA) and transfer RNA (tRNA) interactions within the decoding and translocation centers of the small and large ribosomal subunit.
On one of our studies, we investigated Viomycin which is an antibiotic that has been used to fight tuberculosis infections and been believed to block the translocation step of protein synthesis by inhibiting ribosomal subunit dissociation and trapping the ribosome in an intermediate state of intersubunit rotation. The mechanism by which viomycin stabilizes this state remains unexplained. To address this, we have determined cryoEM and X-ray crystal structures of Escherichia coli 70S ribosome complexes trapped in a rotated state by viomycin. The 3.8-Å resolution cryo-EM structure reveals a ribosome trapped in the hybrid state with 8.6° intersubunit rotation and 5.3° rotation of the 30S subunit head domain, bearing a single P/E state transfer RNA (tRNA). We identify five different binding sites for viomycin, four of which have not been previously described. To resolve the details of their binding interactions, we solved the 3.1-Å crystal structure of a viomycin-bound ribosome complex, revealing that all five viomycins bind to ribosomal RNA. One of these (Vio1) corresponds to the single viomycin that was previously identified in a complex with a nonrotated classical-state ribosome. Three of the newly observed binding sites (Vio3, Vio4, and Vio5) are clustered at intersubunit bridges, consistent with the ability of viomycin to inhibit subunit dissociation. We propose that one or more of these same three viomycins induce intersubunit rotation by selectively binding the rotated state of the ribosome at dynamic elements of 16S and 23S rRNA, thus, blocking conformational changes associated with molecular movements that are required for translocation (Zhang et.al., PNAS, 2020).
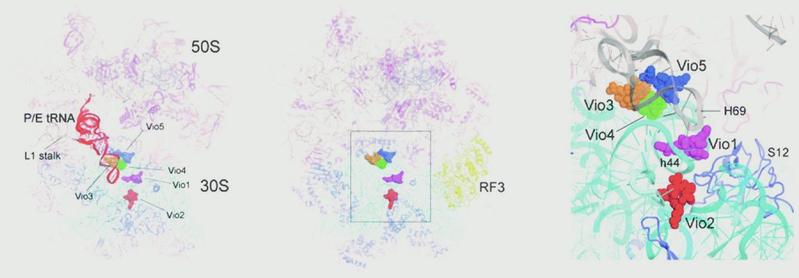
Structures of viomycin-containing complexes
Left: Cryo-EM structure of the E. coli 70S ribosome in a complex with viomycin, mRNA, and a single P/E tRNA (red) in the hybrid state. The positions of the five bound viomycin molecules are indicated. Middle and right: X-ray crystal structure of a viomycin-containing complex of the E. coli 70S ribosome bound with release factor RF3 (yellow), showing the positions of the five viomycin molecules and their surroundings in the ribosome, including 16S rRNA (cyan), 23S rRNA (gray), and ribosomal protein S12 (blue).
Translation regulation
Ribosome translation can be roughly divided into four phases, which are initiation, elongation, termination, and ribosome recycling.
I previously solve a number of bacteria ribosome translocation complexes trapped by elongation factor EF-G (Zhou et.al., Science, 2013, Zhou et.al., Science, 2014). One of our future goals is to expand our studies of translocation into other kingdoms of life, such as study of translocation complexes from eukarya and archaea.